Haplogroup/Haplotree K1a4a1
The following information came from my 23 and Me Personal Page. It gives us an idea of our K1a4a1 haplogroup migration throughout our antroplogical migrational history over the past 180,000 years.
Your haplogroup is determined by your mitochondrial DNA.
Each generation, mothers pass down copies of their mitochondrial DNA (mtDNA) to their children. While most of your genome exists in 23 pairs of chromosomes that exchange pieces between generations in a process called recombination, mtDNA is transmitted unshuffled. Because of this unusual pattern of inheritance, mtDNA contains rich information about maternal lineages.
A small number of DNA changes, called mutations, generally occur from one generation to the next. Because mtDNA does not recombine between generations, these mutations accumulate in patterns that uniquely mark individual lineages. Scientists can compare the sequence differences that result by constructing a tree. This tree shows how maternal lineages relate to one another, including the observation that they all share a most recent common ancestor approximately 150,000 to 200,000 years ago.
The term “haplogroup” refers to a family of lineages that share a common ancestor and, therefore, a particular set of mutations. We identify your haplogroup by determining which branches of the mtDNA tree correspond to your DNA. Because more closely related lineages tend to share geographic roots, your haplogroup can provide insight into the origins of some of your ancient maternal-line ancestors.
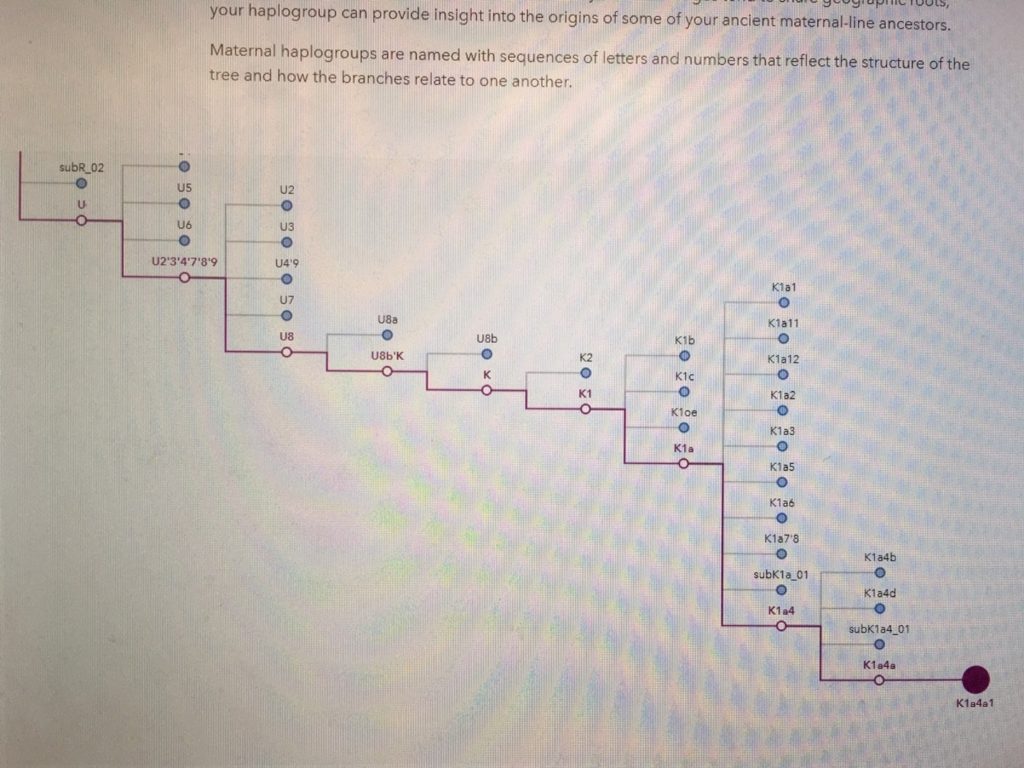
Maternal haplogroups are named with sequences of letters and numbers that reflect the structure of the tree and how the branches relate to one another.
Maternal Haplogroup
You descend from a long line of women that can be traced back to eastern Africa over 150,000 years ago. These are the women of your maternal line, and your maternal haplogroup sheds light on their story.
Mic, your maternal haplogroup is K1a4a1.
Note by Mic -This essay was written by 23 and Me. Family Tree DNA has extended our haplogroup to K1a4a1b2 which is a little further up the Haplotree and closer to present time.
As our ancestors ventured out of eastern Africa, they branched off in diverse groups that crossed and recrossed the globe over tens of thousands of years. Some of their migrations can be traced through haplogroups, families of lineages that descend from a common ancestor. Your maternal haplogroup can reveal the path followed by the women of your maternal line.
Migrations of Your Maternal Line
L 180,000 Years Ago
L3 65,000 Years Ago
N 59,000 Years Ago
R 57,000 Years Ago
U 47,000 Years Ago
K 27,000 Years Ago
Haplogroup L 180,000 Years Ago
If every person living today could trace his or her maternal line back over thousands of generations, all of our lines would meet at a single woman who lived in eastern Africa between 150,000 and 200,000 years ago. Though she was one of perhaps thousands of women alive at the time, only the diverse branches of her haplogroup have survived to today. The story of your maternal line begins with her.
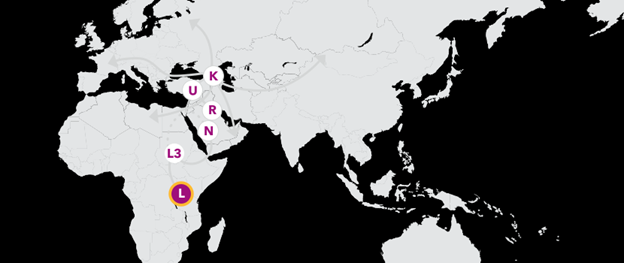
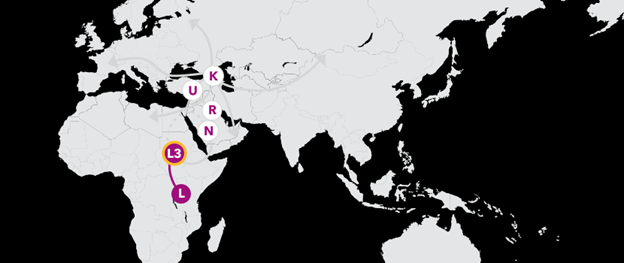
Haplogroup L3 65,000 Years Ago
Your branch of L is haplogroup L3, which arose from a woman who likely lived in eastern Africa between 60,000 and 70,000 years ago. While many of her descendants remained in Africa, one small group ventured east across the Red Sea, likely across the narrow Bab-el-Mandeb into the tip of the Arabian Peninsula.
Haplogroup N 59,000 Years Ago
Your story continues with haplogroup N, one of two branches that arose from L3 in southwestern Asia. Researchers have long debated whether they arrived there via the Sinai Peninsula, or made the hop across the Red Sea at the Bab-el-Mandeb. Though their exact routes are disputed, there is no doubt that the women of haplogroup N migrated across all of Eurasia, giving rise to new branches from Portugal to Polynesia.
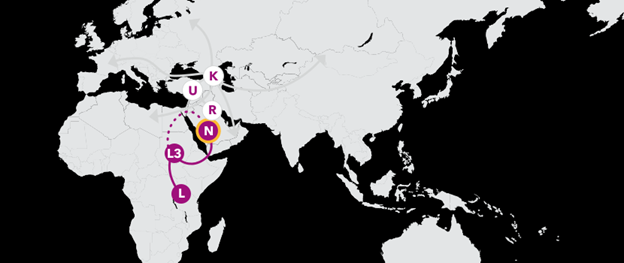
Haplogroup R 57,000 Years Ago
One of those branches is haplogroup R, which traces back to a woman who lived soon after the migration out of Africa. She likely lived in southwest Asia, perhaps in the Arabian peninsula, and her descendants lived and migrated alongside members of haplogroup N. Along the way, R gave rise to a number of branches that are major haplogroups in their own right.
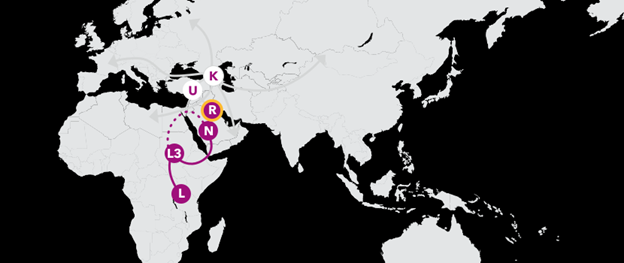
Haplogroup U 47,000 Years Ago
Haplogroup U was one of the earliest offshoots of R, and traces back to a woman who lived nearly 50,000 years ago. Over time, her descendants have migrated into Europe, parts of Asia, and even back into Africa, giving rise to numerous branches spanning the three continents.
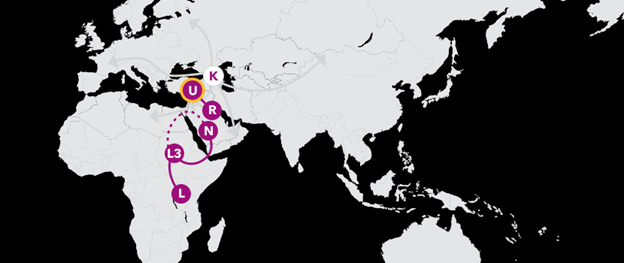
Haplogroup K 27,000 Years Ago
Your maternal-line story continues with haplogroup K, which is actually a branch of haplogroup U8. K traces back to a woman who likely lived in the Middle East less than 30,000 years ago. Since then, her descendants have migrated in all directions, and can be found throughout Europe, Northern Africa, the Arabian Peninsula, and to the east in Central Asia.
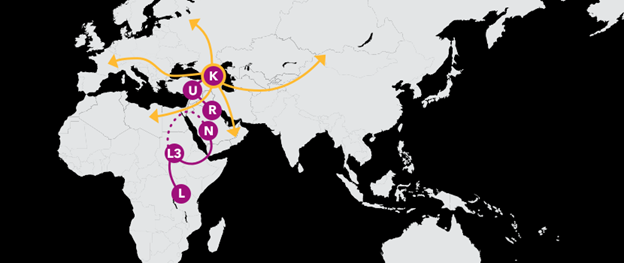
K1a4a1 9,500 Years Ago
Origin and Migrations of Haplogroup K1a4a1
Your maternal line stems from a branch of haplogroup K called K1a4a1. Haplogroup K1a4a1 is predominantly European today, and traces back to a woman who lived approximately 10,000 years ago. Europe was at the start of an age of transformation. To the east in the Fertile Crescent, people were successfully domesticating crops and transitioning to a more settled lifestyle. This new technology supported a boom in human populations, and over thousands of years migrations of these new farmers moved gradually westward across the European continent, leading to additional population growth and great cultural changes. K1a4a1 expanded along with these population changes, giving rise to additional branches throughout Europe.
Today, members of K1a4a1 can be found at low levels within Europe, including in eastern Europe and the Caucasus. Though not typically found outside of Europe, one lineage has been identified as far south as Anatolia.
K1a4a1 Today K1a4a1 is frequent among 23andMe customers.
Today, you share your haplogroup with all the maternal-line descendants of the common ancestor of K1a4a1, including other 23andMe customers.
1 in 190 23andMe customers share your haplogroup assignment.
See references
References
- Abu-Amero KK et al. (2007). “Eurasian and African mitochondrial DNA influences in the Saudi Arabian population.” BMC Evol Biol. 7:32.
- Achilli A et al. (2007). “Mitochondrial DNA variation of modern Tuscans supports the near eastern origin of Etruscans.” Am J Hum Genet. 80(4):759-68.
- Behar DM et al. (2012). “A “Copernican” reassessment of the human mitochondrial DNA tree from its root.” Am J Hum Genet. 90(4):675-84.
- Bodner M et al. (2012). “Rapid coastal spread of First Americans: novel insights from South America’s Southern Cone mitochondrial genomes.” Genome Res. 22(5):811-20.
- Chan EK et al. (2015). “Revised timeline and distribution of the earliest diverged human maternal lineages in southern Africa.” PLoS One. 10(3):e0121223.
- Derbeneva OA et al. (2002). “Traces of early Eurasians in the Mansi of northwest Siberia revealed by mitochondrial DNA analysis.” Am J Hum Genet. 70(4):1009-14.
- Fadhlaoui-Zid K et al. (2004). “Mitochondrial DNA heterogeneity in Tunisian Berbers.” Ann Hum Genet. 68(Pt 3):222-33.
- Fernandes V et al. (2012). “The Arabian cradle: mitochondrial relicts of the first steps along the southern route out of Africa.” Am J Hum Genet. 90(2):347-55.
- Fu Q et al. (2016). “The genetic history of Ice Age Europe.” Nature. 534(7606):200-5.
- González AM et al. (2003). “Mitochondrial DNA affinities at the Atlantic fringe of Europe.” Am J Phys Anthropol. 120(4):391-404.
- Kayser M. (2010). “The human genetic history of Oceania: near and remote views of dispersal.” Curr Biol. 20(4):R194-201.
- Kivisild T et al. (1999). “Deep common ancestry of indian and western-Eurasian mitochondrial DNA lineages.” Curr Biol. 9(22):1331-4.
- Kivisild T et al. (2004). “Ethiopian mitochondrial DNA heritage: tracking gene flow across and around the gate of tears.” Am J Hum Genet. 75(5):752-70.
- López S et al. (2015). “Human Dispersal Out of Africa: A Lasting Debate.” Evol Bioinform Online. 11(Suppl 2):57-68.
- Llamas B et al. (2016). “Ancient mitochondrial DNA provides high-resolution time scale of the peopling of the Americas.” Sci Adv. 2(4):e1501385.
- Majumder PP. (2010). “The human genetic history of South Asia.” Curr Biol. 20(4):R184-7.
- Malaspinas AS et al. (2016). “A genomic history of Aboriginal Australia.” Nature. 538(7624):207-214.
- Malyarchuk BA et al. (2003). “Mitochondrial DNA variability in Bosnians and Slovenians.” Ann Hum Genet. 67(Pt 5):412-25.
- Malyarchuk BA et al. (2006). “Mitochondrial DNA diversity in the Polish Roma.” Ann Hum Genet. 70(Pt 2):195-206.
- Malyarchuk BA et al. (2006). “Mitochondrial DNA variability in the Czech population, with application to the ethnic history of Slavs.” Hum Biol. 78(6):681-96.
- Mellars P et al. (2013). “Genetic and archaeological perspectives on the initial modern human colonization of southern Asia.” Proc Natl Acad Sci U S A. 110(26):10699-704.
- Metspalu M et al. (2004). “Most of the extant mtDNA boundaries in south and southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans.” BMC Genet. 5:26.
- Pereira L et al. (2000). “Diversity of mtDNA lineages in Portugal: not a genetic edge of European variation.” Ann Hum Genet. 64(Pt 6):491-506.
- Plaza S et al. (2003). “Joining the pillars of Hercules: mtDNA sequences show multidirectional gene flow in the western Mediterranean.” Ann Hum Genet. 67(Pt 4):312-28.
- Quintana-Murci L et al. (2004). “Where west meets east: the complex mtDNA landscape of the southwest and Central Asian corridor.” Am J Hum Genet. 74(5):827-45.
- Richard C et al. (2007). “An mtDNA perspective of French genetic variation.” Ann Hum Biol. 34(1):68-79.
- Richards M et al. (2000). “Tracing European founder lineages in the Near Eastern mtDNA pool.” Am J Hum Genet. 67(5):1251-76.
- Richards MB et al. (1998). “Phylogeography of mitochondrial DNA in western Europe.” Ann Hum Genet. 62(Pt 3):241-60.
- Rito T et al. (2013). “The first modern human dispersals across Africa.” PLoS One. 8(11):e80031.
- Sampietro ML et al. (2005). “The genetics of the pre-Roman Iberian Peninsula: a mtDNA study of ancient Iberians.” Ann Hum Genet. 69(Pt 5):535-48.
- Sampietro ML et al. (2007). “Palaeogenetic evidence supports a dual model of Neolithic spreading into Europe.” Proc Biol Sci. 274(1622):2161-7.
- Simoni L et al. (2000). “Geographic patterns of mtDNA diversity in Europe.” Am J Hum Genet. 66(1):262-78.
- Soares P et al. (2009). “Correcting for purifying selection: an improved human mitochondrial molecular clock.” Am J Hum Genet. 84(6):740-59.
- Starikovskaya EB et al. (2005). “Mitochondrial DNA diversity in indigenous populations of the southern extent of Siberia, and the origins of Native American haplogroups.” Ann Hum Genet. 69(Pt 1):67-89.
- Stoneking M et al. (2010). “The human genetic history of East Asia: weaving a complex tapestry.” Curr Biol. 20(4):R188-93.
- Töpf AL et al. (2006). “Tracing the phylogeography of human populations in Britain based on 4th-11th century mtDNA genotypes.” Mol Biol Evol. 23(1):152-61.
- Töpf AL et al. (2007). “Ancient human mtDNA genotypes from England reveal lost variation over the last millennium.” Biol Lett. 3(5):550-3.
- Thanseem I et al. (2006). “Genetic affinities among the lower castes and tribal groups of India: inference from Y chromosome and mitochondrial DNA.” BMC Genet. 7:42.
- Torroni A et al. (1998). “mtDNA analysis reveals a major late Paleolithic population expansion from southwestern to northeastern Europe.” Am J Hum Genet. 62(5):1137-52.
- Yao YG et al. (2004). “Different matrilineal contributions to genetic structure of ethnic groups in the silk road region in china.” Mol Biol Evol. 21(12):2265-80.
